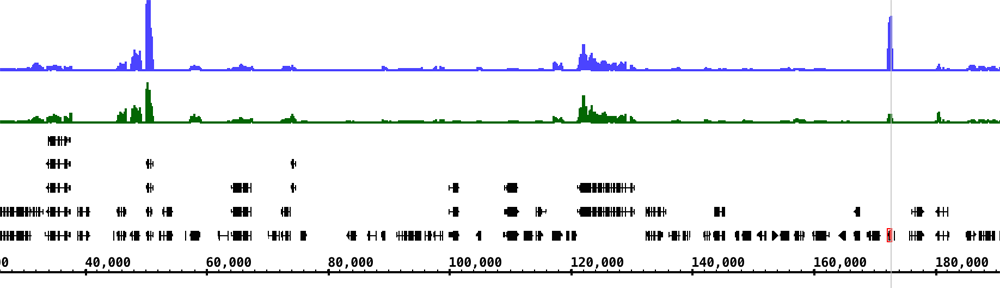
Recently, a researcher working with the sorghum genome posted this question on BioStars – Question: IGB- genome sequence in sequence viewer just shows dashes
Updating to a more recent version of IGB solved the problem, but the question reminded me that Phytozome had released a new sorghum assembly and that we needed to update the IGBQuickLoad site.
In this post, I’ll describe how I updated IGB to the latest sorghum genome assembly using data from Phytozome and a few easy-to-use command line tools.
Step One. Get the data
To start, Nowlan downloaded gene annotations, sequence, and a gene information file from Phytozome.net. The files were:
- Sbicolor_255_v2.0.fa.gz – sequence data
- Sbicolor_255_v2.1.gene_exons.gff3.gz – gene models
- Sbicolor_255_v2.1.defline.txt – gene information file, summarize function
He put them in our shared IGB Dropbox, saving me the trouble of logging in and figuring out where to look. Thank you Nowlan!
Step Two. Find out when the genome was released and what it’s called.
Using Google searches, I found a page describing the new assembly and associated gene annotations. The genome assembly is called “version 2.0” and the annotations are called “version 2.1.”
A search for “Sorghum 2.0 released” found this news page which contained text:
[07 Jun 2013] v2.1 of Sorghum bicolor now available as early release.
This new item refers to the genome annotations, not the genome assembly, but probably the two were released about the same time, so I decided to use this. Why does it matter? It matters because including the month and year in the name of a genome release tells us which release is most recent. This is why IGB gives every assembly a name that tells it (and us) how old the genome is.
Step Three: Check out a copy of IGBQuickLoad.
We are using subversion (a version control system) to track and distribute files that are part of the IGBQuickLoad site.
To add the new data to IGBQuickLoad, I first have to check out a copy using svn. (For more information about svn, see: http://svnbook.red-bean.com/.)
svn co https://svn.transvar.org/repos/genomes/trunk/pub/quickload
I’ve configured my computer to provide the correct user name and password when I execute svn commands. Otherwise, I would need to supply my user name and password uisng the –username and –password options. For more information, run
svn co -h
Step Four: Add new directory for this new genome
Following IGB conventions, I added a new directory to the IGBQuickLoad data repository using svn:
svn mkdir S_bicolor_Jun_2013
By convention, we keep genome assembly data files and meta-data files in a directory named after the species and release date.
I also added new lines to the .htaccess and contents.txt files:
htaccess:
AddDescription "Sorghum bicolor v. 2.0" S_bicolor_Jun_2013
This ensures the directory listing on the IGBQuickLoad.org Web site will report the version name and species.
contents.txt:
S_bicolor_Jun_2013 Sorghum bicolor v2.0 (June 2013)
This ensures IGB will list the genome in the Current Genome tab. When IGB first accesses an IGBQuickLoad site, it fetches this “contents.txt” file and uses it to determine the names and locations of genome assemblies available on the site.
This contents.txt file is a tab-delimited file with two columns – the first column lists genome directories and the second column lists the human-friendly name of the genome, which is what IGB displays in the IGB window title bar when users selects that genome. Note that sometimes two sites could contain different text in the second genome, but this won’t break IGB. IGB will just show the data from whichever file it read first. So far, this has not posed any problems.
Step Five. Make 2bit sequence file.
I used Jim Kent’s faToTwoBit program to convert the fasta sequence file to 2bit. This format is more compact than fasta and is also random access, which means IGB can use the file to retrieve small parts of the genome quickly when users click the Load Sequence button. By convention, we name the 2bit file after the genome version – this ensures IGB will be able to find it on the site.
faToTwoBit Sbicolor_255_v2.0.fa S_bicolor_Jun_2013.2bit
The fasta file is 702 Mb (almost 1 Gb!) and the 2bit file was only 173M. Much better!
This file must reside in the genome directory S_bicolor_Jun_2013. Because it is not very big, I used svn to add it to the repository.
Step Six. Make genome.txt file.
To populate the Current Sequence tab, IGB needs a “genome.txt” file that lists chromosomes and their sizes – this file, like the 2bit file, resides the in the genome directory. To make this, I use twoBitInfo, another Jim Kent program:
twoBitInfo S_bicolor_Jun_2013.2bit genome.txt
IGB’s Current Sequence table will list the chromosomes in the same order they appear in this file, so I view the first few lines of the file to make sure the most useful (fully assembled) chromosomes appear first in the list:
Chr01 73727935
Chr02 77694824
Chr03 74408397
Chr04 67966759
Chr05 62243505
Chr06 62192017
Chr07 64263908
Chr08 55354556
Chr09 59454246
Chr10 61085274
super_10 8818317
This looks fine. But if it didn’t I might re-order the listing by sequence size:
sort -k2,2nr genome.txt > tmp
mv tmp genome.txt
Step Seven. Make annotations meta-data file (annots.xml)
Here, I used a program named phytozomeGff3ToBed.py, available from a Loraine Lab git repository. This program converts GFF to BED12 format:
phytozomeGff3ToBed.py -g Sbicolor_255_v2.1.gene_exons.gff3.gz -b tmp.bed
I used a Unix one-liner convert the BED12 file to BED-detail format, sort the file by sequence name and gene model start position, compress it using bgzip, and then save the result to a new file named for the genome version:
bedToBedDetail.py -g Sbicolor_255_v2.1.defline.txt -b tmp.bed | sort -k1,1 -k2,2n | bgzip > S_bicolor_Jun_2013.bed.gz
To share reference gene model annotations, we use BED-detail format. It’s compact and more convenient for data analysis than GFF3. It contains all the usual field in a BED12 file, plus two extra fields. In IGB, our BED-detail files
- report locus names in field 13, useful for grouping alternative transcripts arising from the same transcriptional unit
- don’t use the itemRGB field (we don’t color-code gene models, but we might do that someday)
- insert the start position in the thickStart and thickStop fields when a gene model doesn’t encode a protein
When we distribute annotations file, we also index them using tabix, which requires that the indexed file be sorted and block compressed using bgzip. (Google “tabix” to find a copy – it’s open source and associated with the samtools package.)
To index the file using tabix, I did this:
tabix -s 1 -b 2 -e 3 S_bicolor_Jun_2013.bed.gz
This created a file named S_bicolor_Jun_2013.bed.gz.tbi (“tbi” for tabix index) which must reside in the same directory as its corresponding data file for IGB (or other programs) to use it. The index file enables IGB to quickly load the file or retrieve just parts of it via byte-range queries.
Next, I viewed the first few lines of the file:
Chr01 24317 38116 Sobic.001G000400.2 0 - 24631 36898 19 590,255,172,228,134,84,291,333,107,126,166,104,161,901,91,291,85,83,3870,683,1377,1661,2230,8581,8772,9179,9608,9929,10147,10436,10651,10922,11953,12118,12497,13040,13412, Sobic.001G000400 similar to PDR-like ABC transporter
Chr01 24317 42528 Sobic.001G000400.1 0 - 24631 42234 20 590,255,172,228,134,84,291,333,107,126,166,104,161,901,91,291,85,83,121,500, 0,683,1377,1661,2230,8581,8772,9179,9608,9929,10147,10436,10651,10922,11953,12118,12497,13040,17387,17711, Sobic.001G000400 similar to PDR-like ABC transporter
Chr01 50023 50488 Sobic.001G000500.1 0 - 50037 50488 54,263, 0,202, Sobic.001G000500 Not Available
Chr01 53429 60786 Sobic.001G000600.1 0 - 60555 60738 204,419, 0,6938, Sobic.001G000600 Not Available
This looked good. Field 4 contained gene model name, field 13 contained locus name, and field 14 contained a description taken from the gene information file (Sbicolor_255_v2.1.defline.txt), but reported “Not Available” when nothing about that gene was listed.
How many gene models have no information?
gunzip -c S_bicolor_Jun_2013.bed.gz | grep -c "Not Available"
14622
gunzip -c S_bicolor_Jun_2013.bed.gz | wc -l
39441
Out of 39,441 gene models, around 14,000 had no functional information. This isn’t great, but it’s not unusual for a newly annotated genome.
Step Eight. Make a HEADER.html file – documentation
Next, I created a HEADER.html file that documents the files and where they came from. When user visit the new genome directory on the web, they’ll see the HEADER file contents displayed along with a listing of the files.
Step Nine. Make an annots.xml file
To ensure that IGB will show the genome, I next needed to add an “annots.xml” meta-data file that reports the data sets that are available for the genome. Because I’m a little lazy, I use the “svn cp” (svn copy) command to make an annots.xml file using the file from a previous genome version.
svn cp ../S_bicolor_Jan_2009/annots.xml .
I used a text editor to update the data for this new genome:
<files>
<file name="S_bicolor_Jun_2013.bed.gz"
title="Genes"
description="Version 2.1 gene models from Phytozome, Nov. 2014."
url="S_bicolor_Jun_2013"
foreground="000000"
name_size="14"
direction_type="arrow"
background="FFFFFF"
load_hint="Whole Sequence"
show2tracks="true"
/>
</files>
What all this means:
- name – name of the file in the file system; IGB assumes it resides in the same directory as the annots.xml file, inside the genome directory S_bicolor_Jun_2013
- title – name IGB gives this data set in the Available Data sets section of the Data Access tab. This is also the name IGB displays in the track label once the data are loaded into a track.
- description – displayed as a tooltip when users position the cursor over the “i” icon next to the data set under Available Data Sets.
- url – if users click on the “i” icon, a Web browser will open to this location relative to the IGBQuickLoad root directory. (http://www.igbquickload.org/quickload/S_bicolor_Jun_2013 in this case.)
- foreground – annotation color
- name_size – font size for the track label
- background – background color for the track
- load_hint – how and when to load the data. “Whole Sequence” means: load all the data at once.
- show2tracks – whether or not to divide the annotations into two tracks – one for minus strand genes and another for plus strand genes
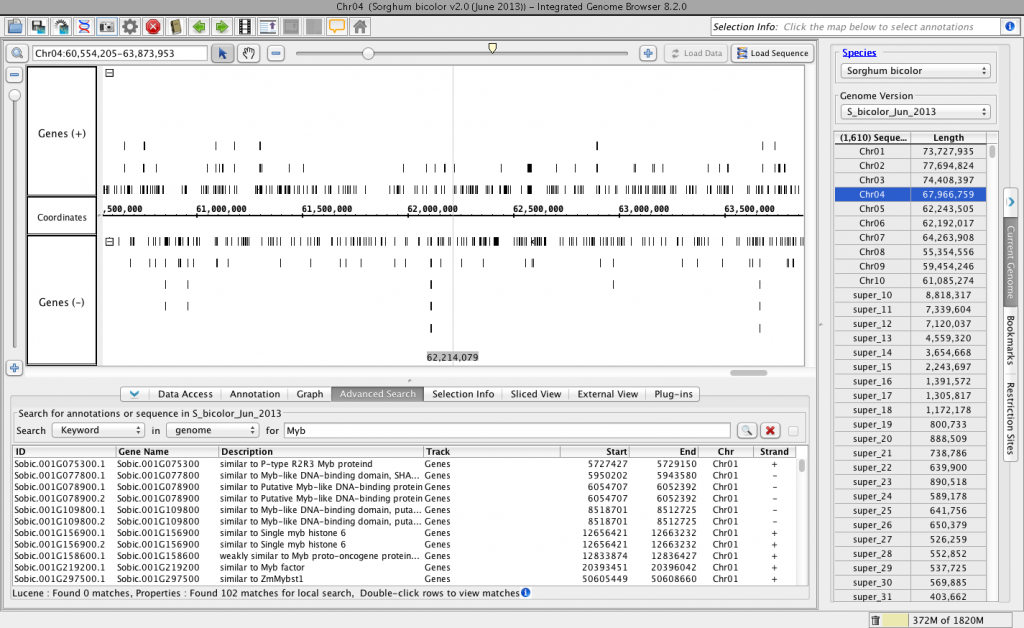
Step Ten. See how it looks in IGB.
For testing and development, I’ve been adding and updating the files in a local copy of the IGBQuickLoad repository that I’ve checked out onto my computer. To see how the new files will look in IGB, I use the Data Sources tab in the Preferences window to add the local QuickLoad site to IGB.
Then, I used the Current Sequence tab to select species Sorghum bicolor and genome version S_bicolor_Jun_2013. I also ran a quick keyword search (Advanced Search tab) to make sure the “details” part of the BED-detail file is read correctly into IGB.
Everything looks good!
Step Eleven. Check it into the repository and update the public IGB QuickLoad sites.
I then used “svn add” and “svn commit” to add the new files to the repository. Then I logged into the main IGBQuickLoad site and the IGBquickLoad backup site and ran “svn up” to add new files and update old ones.
This entire process, including writing this post and coffee breaks, took about two hours of my time.