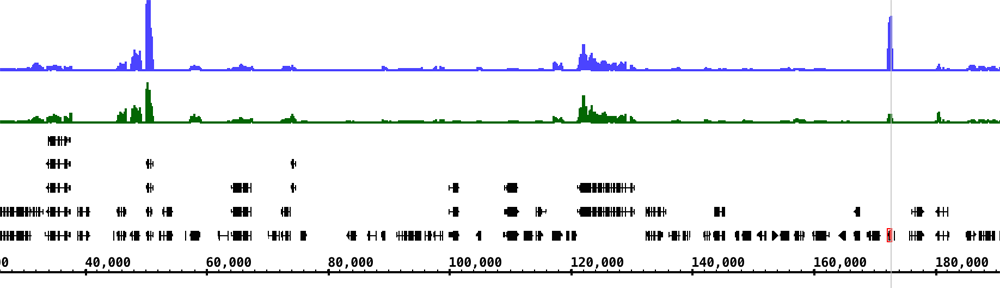
The UCSC Genome Bioinformatics site offers a wealth of data from many genomes, ranging from tiny (but deadly) genomes like ebola virus to much larger genomes like our own human genome. Indeed, the reference genome sequence and gene model data sets for many of the genomes the IGB teams hosts on the IGBQuickLoad.org data repository site are originally from UCSC. Other projects – like Galaxy – also rely on UCSC for core data sets. If you have used Galaxy, you may have noticed that Galaxy has built-in genomes for many animal species; these data were imported from the UCSC ecosystem.
When you visit a UCSC-supported genome in IGB, you’ll see a folder named “UCSC (DAS)” in the Available Data Sets section of the Data Access Panel. If you open the folder, you’ll see many data sets with seemingly cryptic titles – like “nestedRepeats” and “altLocations.” These names correspond to tables in the UCSC database. If you select these data sets and click Load Data in IGB, these data will flow from the UCSC system’s Distributed Annotation Server into IGB.
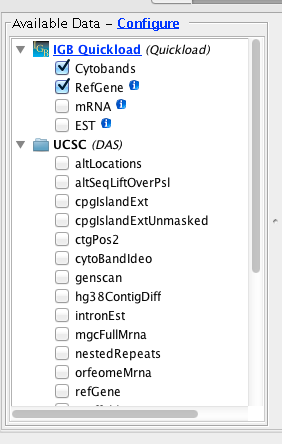 However, for technical reasons, the UCSC DAS site can only support some of their data sets. To view UCSC data that are not supported in their DAS site, you can use the UCSC Table Browser to download them onto your computer and open them in IGB.
However, for technical reasons, the UCSC DAS site can only support some of their data sets. To view UCSC data that are not supported in their DAS site, you can use the UCSC Table Browser to download them onto your computer and open them in IGB.
In this post, I’ll explain how you can use the UCSC Table Browser to obtain and then open data sets in Integrated Genome Browser. I’ll also explain how you can use tabix and bgzip to compress and index files that are too large to load into IGB all at once.
Part I: Getting data from the UCSC Web site
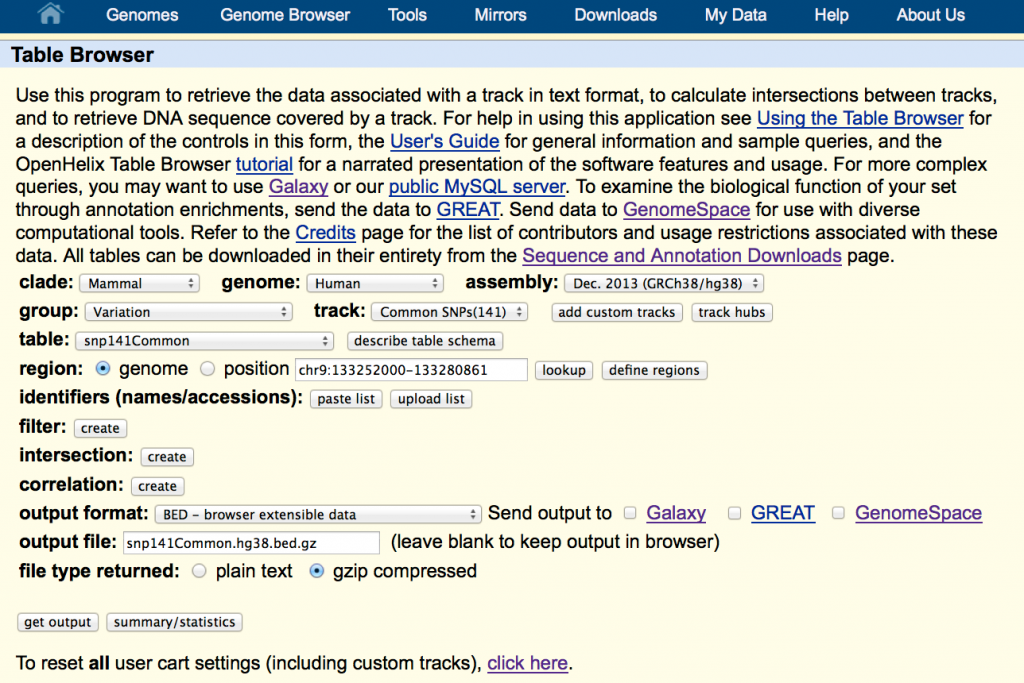
In this example, I’ll show you how to get human variation data (SNPs) from the UCSC Web site.
- Go to http://genome.ucsc.edu
- Click Tables (top of page)
Configure the browser to access the latest (as of this writing) human genome assembly:
- clade Mammal
- genome Human
- assembly Dec 2013 (GRCh38/hg38) (the latest)
- group Variation
- track Common SNPs(141)
- table snp141Common
- region genome
- output format BED – browser extensible data
- file type returned gzip compressed
Enter a name for the output file – e.g., snp141Common.hg38.bed.gz. It should end with “.gz” to ensure your computer will recognize it as a gzip-compressed file.
Tip: Click the button Describe Table Schema to see what data are in the table.
to be continued