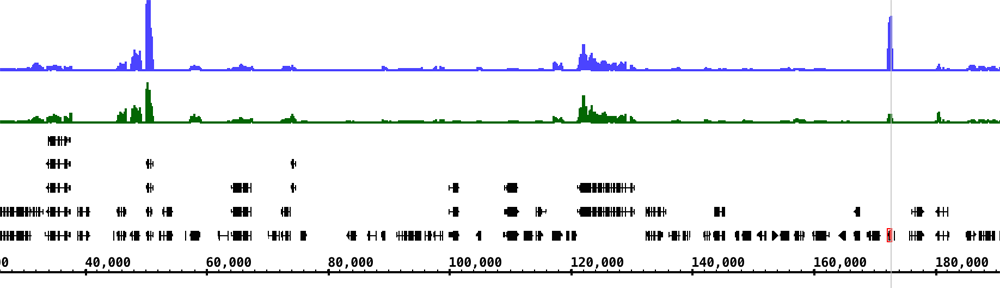
Visualizing genomes
 Scientists need software to explore and analyze genome data. To meet this need, we develop and maintain Integrated Genome Browser, a desktop visualization application (written in Java) used by thousands of researchers worldwide.
Scientists need software to explore and analyze genome data. To meet this need, we develop and maintain Integrated Genome Browser, a desktop visualization application (written in Java) used by thousands of researchers worldwide.
Originally developed in the early 2000s at Affymetrix for visualizing tiling array data, IGB is now open source, free software you can use to visually analyze many other kinds of data, especially data from RNA-Seq, ChIP-Seq, and other *Seq experiments.
Splicing under stress
Plants live their entire lives in one location and must acclimate to daily and seasonal fluctuations in temperature, water availability, and sunlight. Studying how cellular processes in plants adapt to environmental challenges will increase knowledge of how these processes function in all organisms, including humans. It will also help us better understand and counteract negative effects of global warming on agriculture.
We are interested in how plants execute pre-mRNA splicing under different circumstances and developmental stages.
See:
- Alternative splicing analysis code – https://bitbucket.org/lorainelab/altspliceanalysis
- Effects of high temperature and desiccation on splicing in the model plant Arabidopsis thaliana – https://bitbucket.org/lorainelab/hot-dry-arabidopsis
- Alternative splicing in rice – https://bitbucket.org/lorainelab/ricealtsplice
Understanding the genetic basis for heat stress resilience during pollination
Pollination is the process by a which a pollen grain delivers male gametes (sperm) to the female gametes (egg) via tube-like structure called a pollen tube. Once the pollen tube reaches the egg, it bursts, releasing the sperm cells to fuse with the egg and other cells that help form the seed. If fertilization fails, the flower stays a flower forever and never develops into something edible – a grain, fruit, or vegetable. Nearly every kind of plant product we depend on comes from fertilization.
We worked with collaborators at Brown University, Wake Forest University, Arizona State University, and UNC Charlotte to understand how some tomato varieties can withstand high temperatures during pollination while others cannot. Our goal was to identify and document these pathways and mechanisms that help pollination occur at higher-than-usual temperatures.
For more information, see:
- Project summary (National Science Foundation grant abstract): https://www.nsf.gov/awardsearch/showAward?AWD_ID=1939255
- Data processing and analysis code (in progress!): https://bitbucket.org/hotpollen/
Regulation of floral growth and patterning in Arabidopsis thaliana
We worked with Professor Beth Krizek of University of South Carolina to understand how flowers form.
Nearly every plant-based agricultural product – from grains to fruits and vegetables – originate as flowers. Working with Dr. Krizek, we investigated DNA binding proteins that orchestrate flower development in the model plant Arabidopsis thaliana.
For more information, see:
- Project summary (National Science Foundation grant abstract): https://www.nsf.gov/awardsearch/showAward?AWD_ID=1354452
- Data processing and analysis code: https://bitbucket.org/krizeklab/