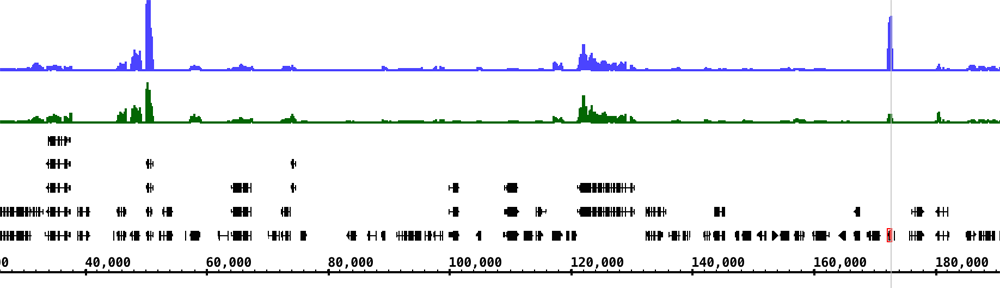
This month, we introduced high school students to the field of personal genomics. Ivory, April, and I taught kids how to analyze genetic variation data from 23&Me using Integrated Genome Browser. Ann attended and worked through the exercises along with the kids.
This week, we showed 4th graders how we grow plants in the lab.


I explained that one of the biggest reasons we study plants is to develop hardier, more nutritious crops. I showed them Arabidopsis plants and introduced the concept of a “model” organism in research. Arabidopsis plants are tiny and grow quickly – like weeds – which makes them ideal for quickly testing theories about how plants grow.
Then, they got their hands dirty transplanting radish seedlings from petri dishes into soil – just like we do with Arabidopsis seedlings when we want uniform growth.


Everyone in the Loraine Lab had a lot of fun helping out with the Scientist for a Day program. I’m sure we all learned as much from the experience as the students. We all look forward to showing of our research to more Kannapolis students next school year.